anderslab
Gonzaga University
Gonzaga University
I am fascinated by the role that DNA plays in living things: how genes contribute to the traits of an organism, how genes are regulated, and how genes and genomes can change from generation to generation. To study genes and genomes, my research makes use of a group of newly-discovered viruses that infect bacteria.
We have been pursuing three kinds of projects:
Mauris placerat lacinia justo vitae feugiat. Vestibulum convallis leo vel tortor ultricies aliquam.
alskj;laksjdf;laskjdf
a;sldfkja;sldfkjasdf
a;sdlkfja;sldfkjas;dlfkj
as;ldfkja;sldfkjas;ldfkj
Check out our latest posts.
Etiam faucibus turpis id ipsum egestas porta. Cras in aliquet purus, ac varius turpis.
Duis consequat ut quam ut sollicitudin. Donec eget congue ligula, eget pharetra urna. Nam tempor tellus sit amet bibendum dapibus.
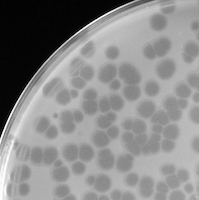
Nam auctor elementum dolor. Donec euismod, justo sed convallis blandit, ipsum erat mattis lectus, vel pharetra neque enim tristique risus.

Quisque tincidunt risus et enim. Vestibulum gravida sem at sem bibendum vehicula. Sed et leo.

Curabitur commodo arcu vel enim mollis consequat. Nulla pharetra tortor vel arcu. In rhoncus fermentum ipsum.

Nam auctor elementum dolor. Donec euismod, justo sed convallis blandit, ipsum erat mattis lectus, vel pharetra neque enim tristique risus.
