BIOCHEMISTRY TOPICS
Myoglobin
Myoglobin: An oxygen binding protein. Tertiary structure of myoglobin. The heme prosthetic group. Ligand binding and saturation curves for ligand binding proteins.
Myoglobin is a monomeric oxygen-binding protein containing a heme prosthetic group. Myoglobin, whose physiological role is to facilitate diffusion in muscle, has a hyperbolic oxygen binding curve. The equation for this curve is readily derived from the expression for the oxygen-binding equilibrium and the definition for fractional saturation.
Myoglobin
An examination of the tertiary structure of myoglobin shows that its secondary structure content consists of mostly α helix , and it is devoid of β sheet. In the tertiary structure, the eight α helices pack together mainly in two layers, with adjacent helices mostly antiparallel and sometimes crossing at fairly sharp angles. The overall shape of the molecule is roughly globular, and the heme prosthetic group (see below) is wedged between two helices forming one of the layers.
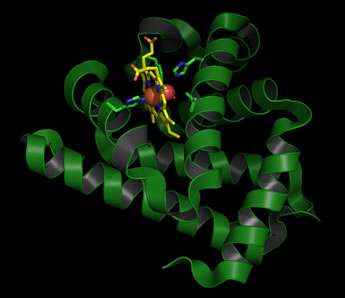
Left: Ribbon diagram of the structure of sperm whale myoglobin (Mb) in its oxygenated (oxy-Mb) form. The heme prothetic group is shown in stick form, with carbon atoms colored yellow, nitrogen blue, and oxygen red. The iron at the center of the heme group is shown as a brick-red sphere, with the adjacent bound O2 molecule as red spheres. Residues from the protein that are involved in iron coordination, or positioning the iron-bound oxygen (the two histidine residues), as well as two additional residues that pack next to the heme are also shown in stick form (carbons are colored green).
Myoglobin often appears in biochemistry textbooks as a classic example of an alpha-helical protein. In fact, Mb was the first protein for which a tertiary structure was determined. This was accomplished for sperm whale Mb in 1959 by John Kendrew using the method of X-ray crystallography - a landmark achievement in the history of biochemistry.
The heme group. The heme prosthetic group of myoglobin provides its oxygen binding site. The central iron ion (Fe2+ in functional Mb) is surrounded by a chemical grouping of four pyrrole rings linked by methene bridges. Such metallated tetrapyrrole ring systems are termed porphyrins. Another important example of a porphyrin is chlorophyll, in which Mg2+ is bound in the middle of the tetrapyrrole ring. In heme, iron receives four coordination bonds from the tetrapyrrole nitrogen atoms in a roughly planar arrangement. Two additional bonds from either side of this plane would complete an octahedral coordination sphere around the iron. In myoglobin, a fifth coordination bond is supplied by a His side chain. The sixth coodination site is also the oxygen binding site.
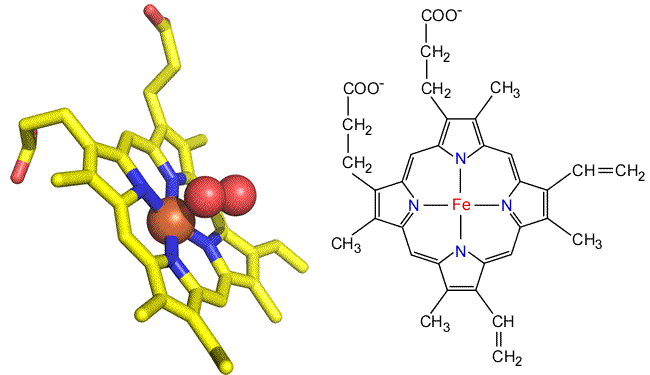
Above: Two views of the heme prosthetic group as found in myoglobin and hemoglobin (heme b or iron protoporphyrin IX). Left: Stick form of oxygenated heme taken from the myoglobin structure. Right: Structural formula of heme b.
Heme occurs in slightly different forms in other heme-containng proteins. The heme found in myoglobin and hemoglobin are b-type hemes, also denoted as iron protoporphyrin IX. In the cytochromes, a family of heme-containing proteins, there are a- and c-type hemes, as well as cytochorome b's. Compared to the iron protoporphyrin IX of b-type hemes, the a-type heme includes a long isoprenoid tail, and a formyl group in place of one of the methyl groups decorating the perimeter of the tetrapyrrole ring nucleus. In c-type hemes, both vinyl groups react with cysteine side chains to covalently link the prosthetic group to the protein, and a Met residue (rather than His) forms an axial ligand to the iron.
The oxygen binding function of myoglobin, as well as that of hemoglobin, requires that the iron remain in the iron(II) or the +2 oxidation state. Oxidation of this iron to +3 destroys oxygen binding capacity; the iron(III) forms of Mb and Hb are called metmyoglobin and methemoglobin, respectively. These functional features of heme in Mb and Hb contrast with the role of iron in the cytochromes, which serve as electron transfer proteins through their ability to alternate between +2 and +3 oxidation states.
Definitions and equations
A ligand is a molecule that is specifically bound by a protein. It can be a small or large molecule, like a a cofactor or coenzyme, or even another protein. A protein together with its ligand is termed the holoprotein, or denoted as "the complex". The protein alone is the apoprotein. In enzymology, the related terms holoenzyme and apoenzyme are commonly used.
Let us represent the ligand as A, the free protein (apoprotein) as P, and the protein-ligand complex (holoprotein) as P·A. The equilibrium between the protein-ligand complex and free ligand plus apoprotein can be represented as
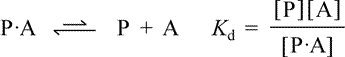
Kd, the equilibrium constant for ligand dissociation, or dissociation constant, is defined in terms of the free ligand concentration, [A], the concentration of the free protein (apoprotein) [P], and the concentration of the protein-ligand complex [P·A]. Rearranging this definition, we see that the ratio of complex to free protein is directly proportional to free ligand concentration, i.e. [A]/Kd = [P·A]/[P].
An experimentally more useful quantity is the fraction Y of protein with the ligand bound, or fractional saturation.
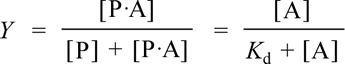
The first equality follows from the literal definition in that it expresses Y as the ratio of protein with bound ligand to the total protein concentration (ligand-bound plus free). The second equality can be obtained by dividing numerator and denominator by [P], then substituting [A]/Kd for [P·A]/[P], then multiplying numerator and denominator by Kd. The second equation is much more useful, as it expresses Y as a function of the fixed parameter Kd, and the single variable [A], the free ligand concentration. Furthermore, since the total protein concentration is nearly always much less than total ligand concentration, free ligand concentration is nearly the same as total ligand concentration. Note that setting the ligand concentration [A] numerically equal to Kd yields Y = 0.5. A plot of Y vs [A] is called a ligand binding curve, or saturation curve. The form of this curve is hyperbolic.
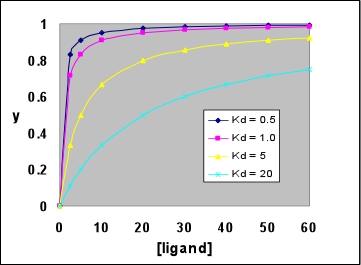
Right: A family of hyperbolic ligand binding curves, according to the above equation, showing the effect of varying Kd. Note that the larger Kd is, the weaker ligand binding to the protein is, and the less saturated the protein is with ligand at any given ligand concentration. In each case, saturation Y reaches 0.5 when the ligand concentration is equal to the Kd value.
Related topics pages: