BIOCHEMISTRY TOPICS
Tertiary structure
Tertiary structure is the overall conformation and resulting three-dimensional shape of a polypeptide chain. Proteins adopt various tertiary structures, ultimately based upon primary structure. Topology diagrams and motifs. Tertiary structure is built up from various combinations of secondary structural features. Example tertiary structures and domains.
Tertiary structure is the overall shape of the polypeptide chain, which depends on the conformation of all the main chain and side chain bonds of the molecule. Experimentally-determined tertiary structures can be explored via publicly-available data and a molecular graphics program or web plug-in. A wealth of protein structure information is readily accessible through the Protein Data Bank (PDB). There are a variety of tools for visualizing and manipulating the structures found there. The PyMOL molecular graphics program is an especially useful and powerful tool, and the images of protein structure representations seen on this website were produced using it.
Proceeding from secondary structure "elements", we can describe how these elements are linked in a particular protein, and the order of β strands in a β sheet structure. This is called protein topology, and can be represented by means of a topology diagram (see figure below). The topology diagram helps us see how the three-dimensional structure of a long polypeptide chain - i.e. protein tertiary structure - can be built up from its secondary structure elements. Topological representation, although abstract, reveals patterns commonly repeated and reiterated throughout structural biology. The recognition of motifs or supersecondary structures as simple "compounds" of secondary structure that bridge the hierarchical gap between secondary and tertiary protein structure can be seen in this light. Given the many thousands of protein structures that have been obtained by the methods of X-ray crystallography and multidimensional NMR, a number of general principles of protein tertiary structure seem well-established.

Left: Example of a topology diagram for a polypeptide chain. The parts of the chain that adopt β strand conformation are shown as yellow arrows, α helices are shown as red cylinders. These secondary structure elements are connected by the blue loops
Motifs and supersecondary structure
Small motifs, like the β-α-β unit introduced earlier, can combine to form larger elements of structure. The figure at left shows two ways in which β-α-β units can combine to form a four-stranded parallel beta sheet. Motifs, supersecondary structure, and domains are terms used to describe structural levels that bridge from simple, repetitive secondary structures to more complex and unique tertiary structures.

The figure at right (click on figure to show large version) shows two ways in which β-α-β units can combine to form a four-stranded parallel β sheet. In the figure, the β-α-β motifs are labeled 1 and 2, and the connecting helices are shown as green cylinders. Part (a) shows a sequential parallel sheet (the order of the strands is 1-2-3-4, from left to right, and all helices lie on the same face of the sheet. In part (b), the order of strands is 4-3-1-2, and the helices from motifs 1 and 2 lie on opposing faces of the sheet. In addition, in this case a crevice or cleft forms between motifs (indicated by the green triangle) at the sheet's C-terminal edge, which is often the location of binding sites for small-molecule ligands or substrates.
Common α/β tertiary structures
Continuing to build on the topologies above, the two most common tertiary topologies emerge, the (α/β)8 or TIM barrel, and the Rossman or nucleotide binding domain.

Further examples of tertiary structures
The common α/β structures described above represent just one of four broad classes of tertiary structure based on secondary structure composition (see image below, or visit the CATH website).
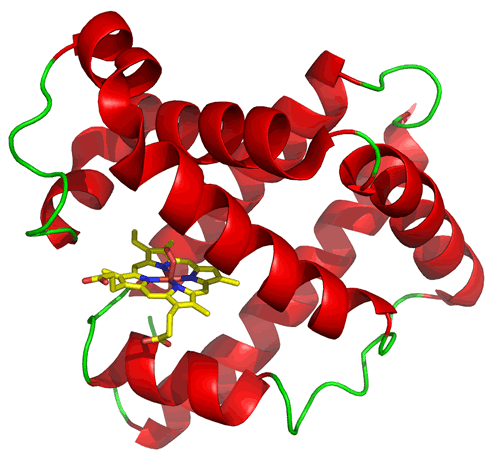
The structure of myoglobin (at left) illustrates a type of tertiary structure composed of α helices (shown in red) connected by loop segments (shown in green). It also provides an example of a common feature in proteins, which is the presence of a non-protein chemical species. In this case, myoglobin has a rather large heme prosthetic group that binds into a cleft or pocket that forms between helices. It is the ironII species at the center of heme that binds oxygen when it is present, which is the function of myoglobin. The protein structure provides the environment that securely binds the heme group and helps main its iron ion in its active +2 oxidation state, thereby maintaining and enhancing its oxygen-binding function.
Domains
In many cases, tertiary structures (such as myoglobin, above) appear to be a single, compact and roughly globular shape. Polypeptide chains between about 40 and 200 residues in length can take on such compact shapes, which average around 25 Å in diameter (1 Å = 1 × 10−10 m = 0.1 nm), and these compact tertiary structures are referred to as domains. Longer polypeptide chains commonly adopt tertiary structures composed of two or more domains. Thus, domains can be thought of as structural units that give the appearance of being capable of existing as separate stable tertiary structures. Very large tertiary structures often form from the folding of long (many hundreds of residues) into multiple domains.

The polypeptide chain that makes up the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) adopts a tertiary structure that appears composed of two separate portions, or domains, which in this case correspond to the first ~150 residues (the lower portion, shown in cyan in the figure), and the following ~185 residues (the upper, tan-colored portion). The N-terminal domain is based upon the Rossman fold architecture built up from α/β motifs described above, forming an extensive parallel β sheet flanked on either side by α helices. This domain is shown binding the dinucleotide redox cofactor NAD+. The C-terminal domain, which contains a number of residues crucial to the catalytic mechanism of GAPDH, is also composed of both α and β secondary structures, but its six-stranded α/β sheet is antiparallel, with α helices arranged on one side of the sheet, between it and the N-terminal domain. A common feature of enzymes with such a two-domain tertiary structure is that a cleft formed between the two domains constitutes a substrate binding pocket and the is location of the enzyme active site.
General features of protein tertiary structure. We can summarize much of what has been learned about protein structure in the form of some generalizations that describe features common to most if not all proteins.
- Proteins have well-packed, non-polar interiors, whereas polar and charged groups are accessible to the outside. Where polar groups occur in the interior (e.g. main chain carbonyls and amides), they tend to be paired.
- The interior of proteins are close-packed, i.e. there are no large cavities.
- The protein chains do not generally form "knots" (although see Taylor & Lin (2003).
- In comparing proteins, it is found that sequence (1° structure) similarity usually implies structural similarity. Note that there are numerous instances in which two proteins with little or no detectable sequence similarity nonetheless adopt very similar structures.
- Overall, there is very little strain in protein structures. In cases where local strain exists, the energetic cost is covered by numerous favorable interactions within the rest of the structure.
.gif)
.gif)
Related topics pages: